Many Variant Strains Were Already Present Before the First Known Cases
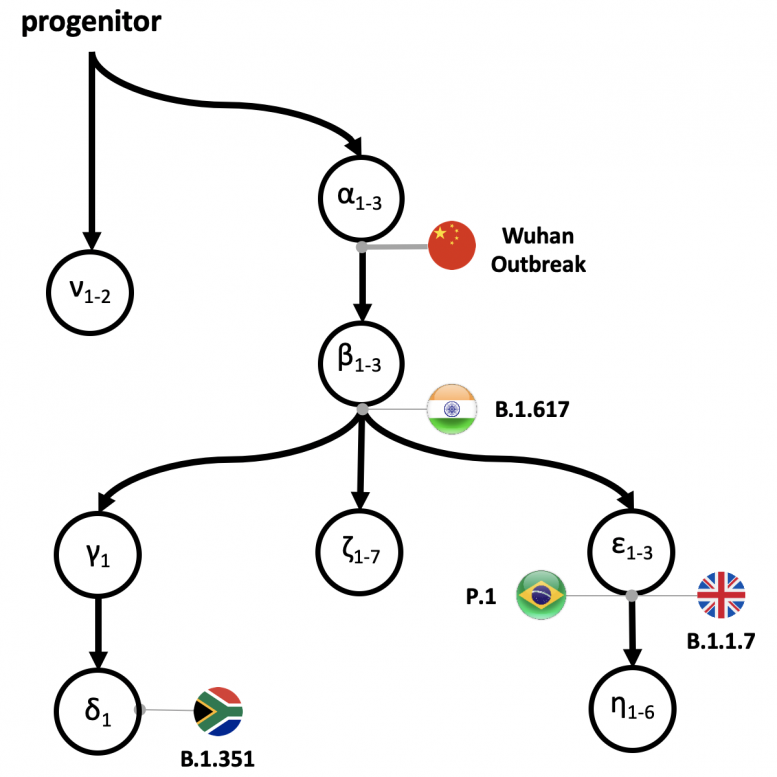
The progenitor (proCoV2) virus and its initial descendants arose in China, based on the earliest mutations of proCoV2 and their locations, which were traced back to occur 6-8 weeks prior to the Wuhan China outbreak. Furthermore, the science team also demonstrated that a population of strains with at least three mutational differences (alpha 1-3) from proCoV2 existed at the time of the first detection of COVID-19 cases in China. The current major variants of interest including the UK (B.1.1.1.7), South African (B.1.351), South American (P.1) and now, Indian (B.1.617) are shown within the pedigree. These variants have not only come to replace prior dominant strains in their respective regions, but still threaten world health due to their potential to escape today’s vaccines and therapeutics. Credit: Sudhir Kumar, Temple University
New study traces back the progenitor genomes causing COVID-19 and geospatial spread.
In the field of molecular epidemiology, the worldwide scientific community has been steadily sleuthing to solve the riddle of the early history of SARS-CoV-2. Despite recent efforts by the World Health Organization, no one to date has identified the first case of human transmission, or “patient zero” in the COVID-19 pandemic.
Finding the earliest possible case is needed to better understand how the virus may have jumped from its animal host first to infect humans as well as the history of how the SARS-CoV-2 viral genome has mutated over time and spread globally.
Since the first SARS-CoV-2 virus infection was detected in December 2019, well over a million genomes of SARS-CoV-2 have been sequenced worldwide, revealing that the coronavirus is mutating, albeit slowly, at a rate of 25 mutations per genome per year. The sheer number of emerging variants, including the UK (B.1.1.1.7), South African (B.1.351), South American (P.1) and now, Indian (B.1.617) have not only come to replace prior dominant strains in their respective regions, but still threaten world health due to their potential to escape today’s vaccines and therapeutics.
“The SARS-CoV-2 virus has already infected more than 145 million people and caused 3 million deaths across the world,” said Sudhir Kumar, director of the Institute for Genomics and Evolutionary Medicine, Temple University. “We set out to find the genetic common ancestor of all these infections, which we call the progenitor genome.”
This progenitor genome (proCoV2) is the mother of all SARS-CoV-2 coronaviruses that has infected and continues to infect people today.
In the absence of patient zero, Kumar and his research team now may have found the next best thing to aid the worldwide molecular epidemiology detective work. “We reconstructed the genome of the progenitor and its early pedigree by using a big dataset of coronavirus genomes obtained from infected individuals since December 2019,” said Kumar, the lead author of a new study, appearing in advanced online edition of the journal Molecular Biology and Evolution.
They found that the progenitor gave rise to a family of coronavirus strains, whose members included the strains found in Wuhan, China, in December 2019. “In essence, the events in December in Wuhan, China, represented the first superspreader event of a virus that had all the tools necessary to cause a worldwide pandemic right out of the box,” said Kumar.
Kumar’s group estimates that the SARS-CoV-2 progenitor was already circulating with an earlier timeline — at least 6 to 8 weeks prior to the first genome sequenced in China, known as Wuhan-1. “This timeline puts the presence of proCoV2 in late October 2019, which is consistent with the report of a fragment of spike protein identical to Wuhan-1 in early December in Italy, among other evidence,” said Sayaka Miura, a senior author of the study.
“We have found progenitor genetic fingerprint in January 2020 and later in multiple coronavirus infections in China and the USA. The progenitor was spreading worldwide months before and after the first reported cases of COVID-19 in China,” said Pond.
Besides their findings on SARS-CoV-2’s early history, Kumar’s group also has developed intuitive mutational fingerprints and Greek symbol classification (ν, α, β, γ, δ, and ε) to simplify the categorization of the major strains, sub-strains and variants infecting an individual or colonizing a global region. This may help scientists better trace and provide context for the order of emergence of new variants.
“Overall, our mutational fingerprinting and nomenclature provide a simple way to glean the ancestry of new variants as compared to phylogenetic designations, e.g., B.1.351 and B.1.1.7,” said Kumar.
For example, an α fingerprint refers to genomes that one or more of the α variants and no other subsequent major variants, and αβ fingerprint refers to genomes that contain all α, at least one β variant, and no other major variants.
“With our tools, we observed…
Read More: Many Variant Strains Were Already Present Before the First Known Cases

